2 LandR Biomass_speciesData Module
2.1 Module Overview
2.1.2 Module summary
LandR Biomass_speciesData (hereafter Biomass_speciesData) downloads and pre-processes species percent (% cover) data layers used by other LandR data modules (e.g., Biomass_borealDataPrep) and by the LandR forest simulation module Biomass_core.
2.1.3 Links to other modules
Biomass_speciesData is intended to be used with any LandR modules that require species % cover raster layers (see examples below). See here for all available modules in the LandR ecosystem and select Biomass_speciesData from the drop-down menu to see potential linkages.
Biomass_borealDataPrep: prepares all parameters and inputs (including initial landscape conditions) that Biomass_core needs to run a realistic simulation. Default values/inputs produced are relevant for boreal forests of Western Canada. Used downstream from Biomass_speciesData;
Biomass_core: core forest dynamics simulation module. Used downstream from Biomass_speciesData.
2.2 Module manual
2.2.1 General functioning
Biomass_speciesData accesses and processes species % cover data
for the parametrisation and initialisation of LandR Biomass_core. This module
ensures 1) that all data use the same geospatial geometries and 2) that these
are correctly re-projected to the study area used for parametrisation
(studyAreaLarge polygon), and 3) attempts to sequentially fill-in and replace
the lowest quality data with higher quality data when several data sources are
used. It’s primary output is a RasterStack of species % cover,
with each layer corresponding to a species.
Currently, the module can access the Canadian National Forest Inventory (NFI)
forest attributes kNN dataset [the default; Beaudoin et al. (2017)], the Common
Attribute Schema for Forest Resource Inventories dataset [CASFRI; Cosco (2011)],
the Ontario Forest Resource Inventory (ONFRI), a dataset specific to Alberta
compiled by Paul Pickell, and other Alberta forest inventory datasets. However,
only the NFI kNN data are freely available and access to the other datasets
must be granted by module developers and data owners, and requires a Google
account. Nevertheless, the module is flexible enough that any user can use it to
process additional datasets, provided that an adequate R function is passed to
the module (see types parameter details in the list of
parameters)
When multiple data sources are used, the module will replace lower quality data
with higher quality data following the order specified in the types parameter.
When multiple species of a given data source are to be grouped, %
cover is summed across species of the same group within each pixel. Please see
the sppEquiv in the list of input objects for
information on how to define species groups.
The module can also exclude species % cover layers if they don’t have a minimum % cover value in at least one pixel. The user should still inspect where species is deemed present (e.g., in how many pixels in total), as it is possible that some datasets only have a few pixels where the species is present, but with reported high % cover. In this case, the user may choose to exclude these species a posteriori. The summary plot automatically shown by Biomass_speciesData can help diagnose whether certain species are present in very few pixels (see Fig. 2.1).
2.2.2 List of input objects
Below is the full list of input objects that Biomass_speciesData requires
(Table 2.2). Of these, the only input
that must be provided (i.e., Biomass_speciesData does not have a default
for) is studyAreaLarge.
Of the inputs in Table 2.2, the following are particularly important and deserve special attention:
studyAreaLarge– the polygon defining the area for which species cover data are desired. It can be larger (but never smaller) that the study area used in the simulation of forest dynamics (i.e.,studyAreaobject in Biomass_core), in which case it should fully cover it.sppEquiv– a table of correspondences between different species naming conventions. This table is used across several LandR modules, including Biomass_core. It is particularly important here because it will determine whether and which species (and their cover layers) are merged. For instance, if the user wishes to simulate a generic Picea spp. that includes, Picea glauca, Picea mariana and Picea engelmannii, they will need to provide these three species names in the data column (e.g.,KNNif obtaining forest attribute kNN data layers from the National Forest Inventory), but the same name (e.g., “Pice_Spp”) in the column chosen for the naming convention used throughout the simulation (defined by thesppEquivColparameter). See Table 2.1 for an example.
| Species | KNN | Boreal | Modelled as |
|---|---|---|---|
| Abies balsamea | Abie_Bal | Abie_Bal | Abies balsamea |
| Abies lasiocarpa | Abie_Las | Abie_Las | Abies lasiocarpa |
| Picea engelmannii x glauca | Pice_Eng_Gla | Pice_Spp | Picea spp. |
| Picea engelmannii x glauca | Pice_Eng_Gla | Pice_Spp | Picea spp. |
| Picea engelmannii | Pice_Eng | Pice_Spp | Picea spp. |
| Picea glauca | Pice_Gla | Pice_Spp | Picea spp. |
| Picea mariana | Pice_Mar | Pice_Spp | Picea spp. |
| Pinus contorta | Pinu_Con | Pinu_Con | Pinus contorta |
| objectName | objectClass | desc | sourceURL |
|---|---|---|---|
| rasterToMatchLarge | RasterLayer |
a raster of studyAreaLarge in the same resolution and projection the simulation’s. Defaults to the using the Canadian Forestry Service, National Forest Inventory, kNN-derived stand biomass map.
|
|
| sppColorVect | character |
A named vector of colors to use for plotting. The names must be in sim$sppEquiv[[sim$sppEquivCol]], and should also contain a color for ‘Mixed’
|
NA |
| sppEquiv | data.table |
table of species equivalencies. See LandR::sppEquivalencies_CA.
|
|
| studyAreaLarge | SpatialPolygonsDataFrame |
Polygon to use as the parametrisation study area. Must be provided by the user. Note that studyAreaLarge is only used for parameter estimation, and can be larger than the actual study area used for LandR simulations (e.g, larger than studyArea in LandR Biomass_core).
|
NA |
| studyAreaReporting | SpatialPolygonsDataFrame |
multipolygon (typically smaller/unbuffered than studyAreaLarge and studyArea in LandR Biomass_core) to use for plotting/reporting. If not provided, will default to studyAreaLarge.
|
NA |
2.2.3 List of parameters
Table 2.3 lists all parameters used in Biomass_speciesData and their detailed information. All these parameters have default values specified in the module’s metadata.
Of these parameters, the following are particularly important:
coverThresh– integer. Defines a minimum % cover value (from 0-100) that the species must have in at least one pixel to be considered present in the study area, otherwise it is excluded from the final stack of species layers (speciesLayers). Note that this will affect what species have data for an eventual simulation and the user will need to adjust simulation parameters accordingly (e.g., species in trait tables will need to match the species inspeciesLayers).types– character. Which % cover data sources are to be used (see General functioning). Several data sources can be passed, in which case the module will overlay the lower quality layers with higher quality ones following the order of data sources intypes. For instance, iftypes == c("KNN", "CASFRI", "ForestInventory"), KNN is assumed to be the lowest quality data set and ForestInventory the highest, hence values in KNN layers are replaced with overlapping values from CASFRI layers and values from KNN and CASFRI layers are replaced with overlapping values of ForestInventory layers.
| paramName | paramClass | default | min | max | paramDesc |
|---|---|---|---|---|---|
| coverThresh | integer | 10 | NA | NA | The minimum % cover a species needs to have (per pixel) in the study area to be considered present |
| dataYear | numeric | 2001 | NA | NA |
Passed to paste0('prepSpeciesLayers_', types) function to fetch data from that year (if applicable). Defaults to 2001 as the default kNN year.
|
| sppEquivCol | character | Boreal | NA | NA |
The column in sim$sppEquiv data.table to group species by and use as a naming convention. If different species in, e.g., the kNN data have the same name in the chosen column, their data are merged into one species by summing their % cover in each raster cell.
|
| types | character | KNN | NA | NA |
The possible data sources. These must correspond to a function named paste0('prepSpeciesLayers_', types). Defaults to ‘KNN’ to get the Canadian Forestry Service, National Forest Inventory, kNN-derived species cover maps from year ‘dataYear’, using the LandR::prepSpeciesLayers_KNN function (see https://open.canada.ca/ data/en/dataset/ec9e2659-1c29-4ddb-87a2-6aced147a990 for details on these data). Other currently available options are ‘ONFRI’, ‘CASFRI’, ‘Pickell’ and ‘ForestInventory’, which attempt to get proprietary data - the user must be granted access first. A custom function can be used to retrieve any data, just as long as it is accessible by the module (e.g., in the global environment) and is named as paste0('prepSpeciesLayers_', types).
|
| vegLeadingProportion | numeric | 0.8 | 0 | 1 | a number that defines whether a species is leading for a given pixel. Only used for plotting. |
| .plotInitialTime | numeric | NA | NA | NA | This describes the simulation time at which the first plot event should occur |
| .plotInterval | numeric | NA | NA | NA | This describes the simulation time interval between plot events |
| .saveInitialTime | numeric | NA | NA | NA | This describes the simulation time at which the first save event should occur |
| .saveInterval | numeric | NA | NA | NA | This describes the simulation time interval between save events |
| .sslVerify | integer | 64 | NA | NA |
Passed to httr::config(ssl_verifypeer = P(sim)$.sslVerify) when downloading KNN (NFI) datasets. Set to 0L if necessary to bypass checking the SSL certificate (this may be necessary when NFI’s website SSL certificate is not correctly configured).
|
| .studyAreaName | character | NA | NA | NA |
Human-readable name for the study area used. If NA, a hash of studyAreaLarge will be used.
|
| .useCache | character | init | NA | NA | Controls cache; caches the init event by default |
| .useParallel | numeric | 16 | NA | NA |
Used in reading csv file with fread. Will be passed to data.table::setDTthreads.
|
2.2.4 List of outputs
The module produces the outputs in Table
2.4, and automatically saves the
processed species cover layers in the output path defined in
getPaths(sim)$outputPath.
| objectName | objectClass | desc |
|---|---|---|
| speciesLayers | RasterStack | biomass percentage raster layers by species in Canada species map |
| treed | data.table | Table with one logical column for each species, indicating whether there were non-zero cover values in each pixel. |
| numTreed | numeric | a named vector with number of pixels with non-zero cover values for each species |
| nonZeroCover | numeric | A single value indicating how many pixels have non-zero cover |
2.2.5 Simulation flow and module events
Biomass_speciesData initialises itself and prepares all inputs provided that
it has internet access to download the raw data layers, or that these layers
have been previously downloaded and stored in the folder specified by
options("reproducible.destinationPath")7.
The module defaults to processing cover data fo all species listed in the
Boreal column of the default sppEquiv input data.table object, for which
there are available % cover layers in the kNN dataset (Table
2.5; see ?LandR::sppEquivalencies_CA for
more information):
| Species | Generic name |
|---|---|
| Abies balsamea | Balsam Fir |
| Abies lasiocarpa | Fir |
| Acer negundo | Boxelder maple |
| Acer pensylvanicum | Striped maple |
| Acer saccharum | Sugar maple |
| Acer spicatum | Mountain maple |
| Acer spp. | Maple |
| Alnus spp | Alder |
| Betula alleghaniensis | Swamp birch |
| Betula papyrifera | Paper birch |
| Betula populifolia | Gray birch |
| Betula spp. | Birch |
| Fagus grandifolia | American beech |
| Fraxinus americana | American ash |
| Fraxinus nigra | Black ash |
| Fraxinus spp. | Ash |
| Larix laricina | Tamarack |
| Larix lyallii | Alpine larch |
| Larix occidentalis | Western larch |
| Larix spp. | Larch |
| Picea engelmannii x glauca | Engelmann’s spruce |
| Picea engelmannii x glauca | Engelmann’s spruce |
| Picea engelmannii | Engelmann’s spruce |
| Picea glauca | White.Spruce |
| Picea mariana | Black.Spruce |
| Picea spp. | Spruce |
| Pinus albicaulis | Whitebark pine |
| Pinus banksiana | Jack pine |
| Pinus contorta | Lodgepole pine |
| Pinus monticola | Western white pine |
| Pinus resinosa | Red pine |
| Pinus spp. | Pine |
| Populus balsamifera v. balsamifera | Balsam poplar |
| Populus trichocarpa | Black cottonwood |
| Populus grandidentata | White poplar |
| Populus spp. | Poplar |
| Populus tremuloides | Trembling poplar |
| Tsuga canadensis | Eastern hemlock |
| Tsuga spp. | Hemlock |
Biomass_speciesData only runs two events, the init event where all species
cover layers are processed and a plotting event (initPlot) that plots the
final layers.
The general flow of Biomass_speciesData processes is:
Download (if necessary) and spatial processing of species cover layers from the first data source listed in the
typesparameter. Spatial processing consists in sub-setting the data to the area defined bystudyAreaLargeand ensuring that the spatial projection and resolution match those ofrasterToMatchLarge. After spatial processing, species layers that have no pixels with values \(\ge\)coverThreshare excluded.If more than one data source is listed in
types, the second set of species cover layers is downloaded and processed as above.The second set of layers is assumed to be the highest quality dataset and used to replaced overlapping pixel values on the first (including for species whose layers may have been initially excluded after applying the
coverThreshfilter).Steps 2 and 3 are repeated for remaining data sources listed in
types.Final layers are saved to disk and plotted (
initPlotevent). A summary of number of pixels with forest cover are calculated (treedandnumTreedoutput objects; see list of outputs).
2.3 Usage example
This module can be run stand-alone, but it only compiles species % cover data into layers used by other modules.
2.3.2 Set up R libraries
options(repos = c(CRAN = "https://cloud.r-project.org"))
tempDir <- tempdir()
pkgPath <- file.path(tempDir, "packages", version$platform, paste0(version$major,
".", strsplit(version$minor, "[.]")[[1]][1]))
dir.create(pkgPath, recursive = TRUE)
.libPaths(pkgPath, include.site = FALSE)
if (!require(Require, lib.loc = pkgPath)) {
remotes::install_github(paste0("PredictiveEcology/", "Require@5c44205bf407f613f53546be652a438ef1248147"),
upgrade = FALSE, force = TRUE)
library(Require, lib.loc = pkgPath)
}
setLinuxBinaryRepo()2.3.3 Get the module and module dependencies
Require(paste0("PredictiveEcology/", "SpaDES.project@6d7de6ee12fc967c7c60de44f1aa3b04e6eeb5db"),
require = FALSE, upgrade = FALSE, standAlone = TRUE)
paths <- list(inputPath = normPath(file.path(tempDir, "inputs")),
cachePath = normPath(file.path(tempDir, "cache")), modulePath = normPath(file.path(tempDir,
"modules")), outputPath = normPath(file.path(tempDir,
"outputs")))
SpaDES.project::getModule(modulePath = paths$modulePath, c("PredictiveEcology/Biomass_speciesData@master"),
overwrite = TRUE)
## make sure all necessary packages are installed:
outs <- SpaDES.project::packagesInModules(modulePath = paths$modulePath)
Require(c(unname(unlist(outs)), "SpaDES"), require = FALSE, standAlone = TRUE)
## load necessary packages
Require(c("SpaDES", "LandR", "reproducible"), upgrade = FALSE,
install = FALSE)2.3.4 Setup simulation
For this demonstration we are using all default parameter values, except
coverThresh , which is lowered to 5%. The species layers (the
major output of interest) are saved automatically, so there is no need to tell
spades what to save using the outputs argument (see
?SpaDES.core::outputs).
We pass the global parameter .plotInitialTime = 1 in the simInitAndSpades
function to activate plotting.
# User may want to set some options -- see
# ?reproducibleOptions -- e.g., often the path to the
# 'inputs' folder will be set outside of project by user:
# options(reproducible.inputPaths =
# 'E:/Data/LandR_related/') # to re-use datasets across
# projects
studyAreaLarge <- Cache(randomStudyArea, size = 1e+07, cacheRepo = paths$cachePath) # cache this so it creates a random one only once on a machine
# Pick the species you want to work with -- here we use the
# naming convention in 'Boreal' column of
# LandR::sppEquivalencies_CA (default)
speciesNameConvention <- "Boreal"
speciesToUse <- c("Pice_Gla", "Popu_Tre", "Pinu_Con")
sppEquiv <- LandR::sppEquivalencies_CA[get(speciesNameConvention) %in%
speciesToUse]
# Assign a colour convention for graphics for each species
sppColorVect <- LandR::sppColors(sppEquiv, speciesNameConvention,
newVals = "Mixed", palette = "Set1")
## Usage example
modules <- list("Biomass_speciesData")
objects <- list(studyAreaLarge = studyAreaLarge, sppEquiv = sppEquiv,
sppColorVect = sppColorVect)
params <- list(Biomass_speciesData = list(coverThresh = 5L))2.3.5 Run module
Note that because this is a data module (i.e., only attempts to prepare data for
the simulation) we are not iterating it and so both the start and end times are
set to 1 here.
opts <- options(reproducible.useCache = TRUE, reproducible.destinationPath = paths$inputPath,
reproducible.useCache)
mySimOut <- simInitAndSpades(times = list(start = 1, end = 1),
modules = modules, parameters = params, objects = objects,
paths = paths, .plotInitialTime = 1)
options(opts)Here are some of outputs of Biomass_speciesData (dominant species) in a randomly generated study area within Canada.
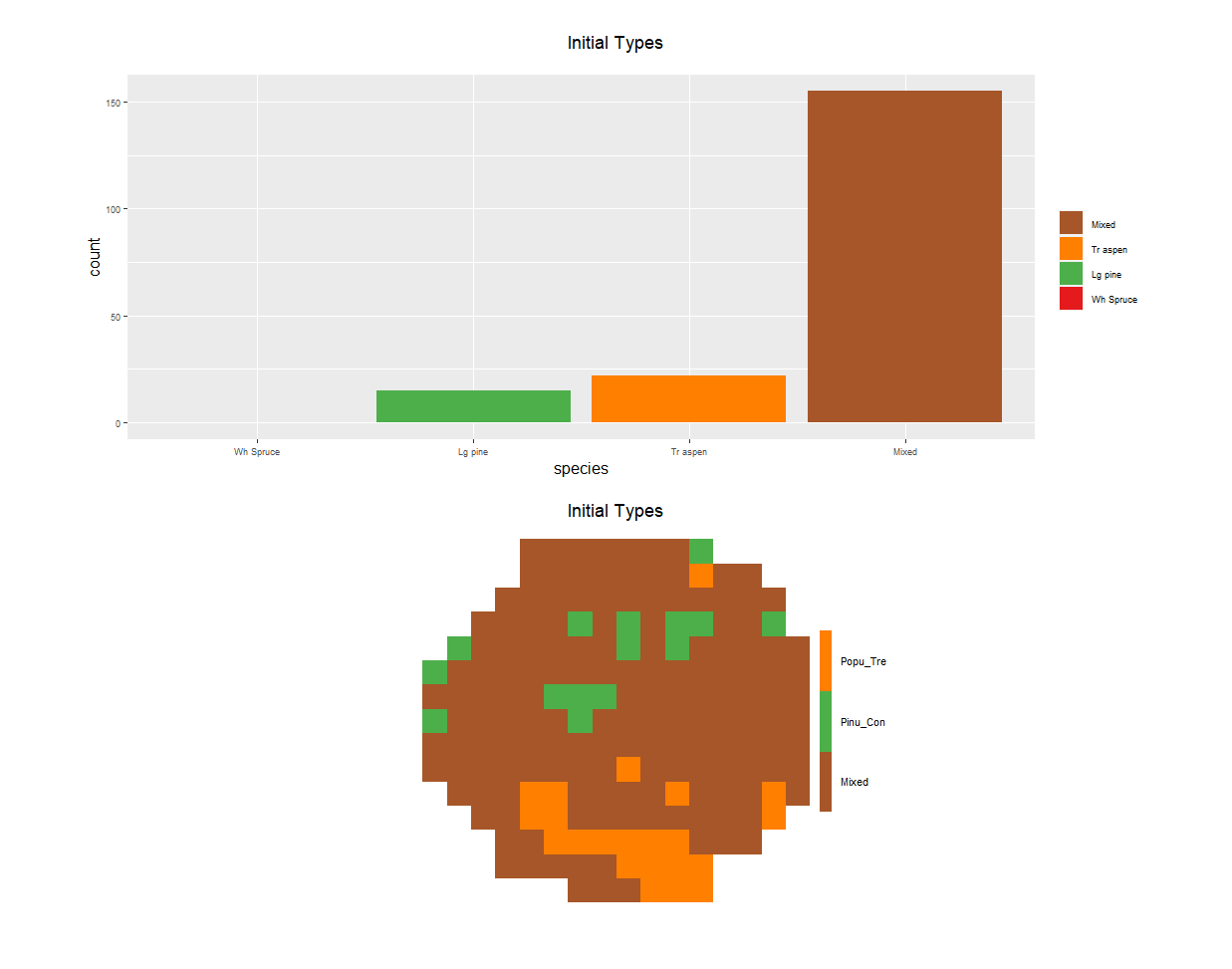
Figure 2.1: Biomass_speciesData automatically generates a plot of species dominance and number of presences in the study area when .plotInitialTime=1 is passed as an argument.
Raw data layers downloaded by the module are saved in `dataPath(sim)`, which can be controlled via `options(reproducible.destinationPath = …)`.↩︎

